[vc_row][vc_column][vc_column_text]
VT News | September 15, 2020
[/vc_column_text][/vc_column][/vc_row][vc_row][vc_column][vc_column_text]
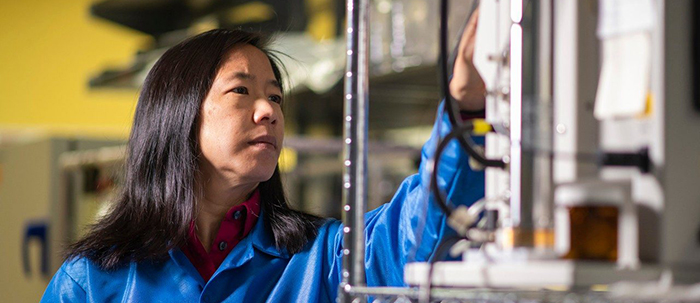
Throughout much of her career, Linsey Marr found that her multi-disciplinary approach to science was more of a challenge than an asset.
“For the past 12 years I feel like I’ve toiled in total obscurity working on airborne viruses,” said Marr, the Charles P. Lunsford Professor of Civil and Environmental Engineering. “It’s an interdisciplinary topic that doesn’t fall neatly into one department or field of study. I’ve struggled at times to get funding and to get papers — except now over the period of a couple of months, it became one of the hottest topics around.”
When the COVID-19 pandemic erupted across the world, and as scientists collected evidence that the virus was transmitted through airborne aerosols, that “total obscurity” suddenly became a bright spotlight. As one of fewer than 12 worldwide experts on aerosol transmission of viruses, and one of only a few in America, Marr’s expertise and ability to communicate to the public became a hot commodity. Since March, she has given more than 300 interviews and been quoted more than 4,000 times in 79 countries.
“Most of us had never heard of aerosol science before the pandemic,” began a New York Times profile headlined, “The Scientist, the Air and the Virus.” “Then Virginia Tech’s Linsey Marr showed up and became our tour guide to the invisible world of airborne particles.”
“It’s been a bit of a spaceship ride,” Marr laughed in an hour-long conversation with Virginia Tech President Tim Sands on Tuesday.
Marr’s interdisciplinary background gives her a broad view of the COVID-19 pandemic, but also the ability to communicate knowledge through memorable analogies and visuals that help her students and the public to better understand the virus and how it moves through the air.
As Sands noted, Marr took an interdisciplinary approach from the beginning of her academic career, “and even broadened rather than narrowed down as you went forward.”
“I hated the idea of having to choose a major,” Marr said. “I loved and appreciated natural science and math and even social science. I tried to pick a major that was as broad as possible.”
Marr holds a B.S. in Engineering Science from Harvard College and a Ph.D. in Civil and Environmental Engineering from the University of California at Berkeley. At Virginia Tech, she leads the Applied Interdisciplinary Research in Air (AIR2) laboratory and teaches courses on air pollution and environmental engineering.
“Virginia Tech has a strong culture of promoting interdisciplinary research,” Marr said. “I felt free to pursue research that was interesting to me. I have great colleagues here. The culture of the university allows you to reach across departments. People are very friendly and open to collaboration. People aren’t trying to protect their turf as much.”
As COVID-19 emerged in Wuhan, China, and began to spread around the world, scientists debated how the virus was transmitted. Given her previous research on the spread of the flu and other diseases, as well as evidence that previous coronaviruses such as SARS spread through aerosols, Marr felt that COVID-19 would spread through aerosols as well.
In early March, Marr posted a thread on Twitter that began, “Let’s talk about #airborne transmission of #SARSCOV2 and other viruses. A discussion is needed to improve accuracy and reduce fear associated with the term.”
Marr told Sands she had posted that thread to bust some myths about airborne viruses and to replace them with more accurate information.
“As soon as people hear ‘airborne virus,’ there’s a tendency to panic,” Marr said. “I knew it was important to get the right information out there. We do know how this moves and there are things we can do to reduce transmission.”
The thread showcased Marr’s ability to communicate simply and effectively, and soon journalists came calling. She became an important voice in the public discourse. Marr used memorable images, describing large water droplets that are expelled by the mouth or nose as “cannonballs.” She compared the airborne transmission of the virus through clouds of microscopic droplets to being with a smoker: If one is indoors and the room is poorly ventilated, the smoke gets pretty thick, whereas if you’re in a car with the windows down, it’s not as bad.
Through the conversation, Sands leveraged Marr’s knowledge and descriptions to essentially walk through a how-to guide for students and the public to navigate this pandemic.
How to wear a mask:
Cover your nose and mouth, with no gaps around the edges.
How do masks work?
Marr described a horse and forest.
“What I like to think about is horse running full speed across a field, and at the edge of the field there’s a forest,” Marr said. “The trees are spaced closely, but they’re far enough apart that if the horse were moving slowly, the horse could get navigate through that forest. But the horse is going full speed, and it hits that the edge of the forest and it’s going to crash into the trees.
So that’s one way that the fibers in your mask trap particles that are coming at them. They [the particles] can’t make the turns that the airflow has to make around those fibers.”
Research shows that layering fabric improves a mask’s effectiveness. A single layer of fabric helps some, but two layers should block 95 percent of particles, Marr said.
How to gauge risk:
Remember the three Cs and the M.
- Avoid Close contact situations. Maintain distance from other people. Six feet doesn’t mark a magic barrier, but the particles do dilute with distance.
- Avoid Crowds. Being in large crowds increase the chances that one will be around someone infected with the virus, and that the virus will spread to others.
- Avoid Closed, poorly ventilated spaces. Imagine a cigarette smoker. In this room, will I take in any smoke?
- Wear Masks.
How to ride in a car:
Roll the windows down, even a few inches, and the air refreshes in a minute or two.
How to ride in an airplane:
The ventilation systems really help and are a point of pride for airlines. However, keep your mask on to reduce the risk of transmission from people in the row with you, in front of, and behind you.
How to socialize when it gets colder:
Marr suggested using blankets to tolerate cold temperatures outside, or indoors when all the windows are open. Another possibility for socializing outside would be to gather around a fire, if everyone is masked and keeping distance.
How to endure flu season:
Marr is optimistic.
“All the interventions that we’re taking for COVID-19 will also be effective for the flu, and so I expect we’ll see one of our mildest flu seasons ever,” Marr said. “The flu transmits largely in the in the same ways that COVID-19 transmits. There’s really a lot similarities, and I think we’ll see a lot fewer flus and other respiratory diseases this coming winter. But we need to remain vigilant.”
How to drink a beverage while wearing a mask:
Marr said she’d considered this at some length. Removing one’s mask to drink doesn’t really work. So she landed on the idea of combining a mask with a bendy straw — though Marr acknowledged with a laugh she hasn’t really tested this idea, either.
[/vc_column_text][/vc_column][/vc_row][vc_row][vc_column][vc_separator][/vc_column][/vc_row][vc_row][vc_column][vc_column_text]
CONTACT:
[/vc_column_text][/vc_column][/vc_row][vc_row][vc_column][vc_separator style=”shadow”][/vc_column][/vc_row]